In some senses, the cell is to biology what the atom is to physics: a basic unit, a fundamental building block of the objects one is studying (living organisms for biologists, material objects in general for physicists). Cells are wonderful creatures: they can do an amazingly wide variety of things. Think of heart cells, which work to pump blood to different parts of the body; or blood cells themselves, which carry oxygen and fight infections; or muscle cells, which allow us to move our arms and legs by stretching and contracting; or brain cells (neurons), which form complicated circuits that carry out all sort of calculations, and ultimately underlie our thoughts, memories and emotions. And yet the amazing thing is that all of these cells contain the same 'recipe book', the sequences of DNA (or genes) that are the 'blueprint for life'. So how is it that cells that all have the same recipe book manage to behave so differently? The answer is that each cell is picking out its favourite recipes (genes) to put into practice. So if a gene is like a recipe, then what is it a recipe for? A protein, of course! Indeed, all cells contain big kitchens, or factories (technically called ribosomes), whose job is to read the recipes and use them to create new proteins. If cells are the building blocks of life, then proteins are the building blocks of cells; all the different parts and bits of machinery inside a cell are largely made of proteins — including the factories that make new proteins! (Think of a brick kiln, made of bricks that were themselves made in another brick kiln.)
Returning to the amazing versatility displayed by cells, we now know that the reason they are so varied is because they are producing different combinations of proteins. Proteins are a bit like lego pieces: they come in many different shapes and sizes, and they can join together to make all sorts of fancy structures that can do useful things. In fact, proteins are better than lego pieces: they can even wiggle about, and fold up in different ways, and change their shape altogether when they attach to something else!
 |
But how exactly does a particular cocktail of proteins lead to a cell with a specific kind of structure and behaviour, such as a muscle or brain cell? We don't really understand this, and it is something many scientists are working on. Figuring out how the different parts fit together and what roles each of them plays is important for all sorts of reasons; for example, it can help to tell us what exactly has gone wrong when cells start to behave badly, as in cancer, when they begin growing too quickly. If we can pin down the rogue protein(s), it makes it much easier to try and design drugs to fix the problem. Unfortunately, biology is very complicated: we are not very good at understanding the structure and behaviour of even a single protein, and when lots of proteins interact and join up in complex ways, it becomes all the more bewildering!
Thankfully, there are techniques for coming up with simple representations of systems that are made up of many interacting parts. One way of doing this is to think of the system as a network. What is a network? It's just what you might expect; like we have computer networks, or railway networks, or Facebook networks, we can also have protein networks:
 |
In the above picture, each dot is a protein, and two dots are joined by a line if we know that the two proteins can join up in some way (just like if two lego blocks can fit into each other). Also, we have coloured the proteins according to which 'community' they belong to. So what's a community? Basically, it's a group of things (computers, people, proteins) that interact a lot more amongst themselves than they do with things outside their group. In the network above, we can clearly see such groups of proteins, and we've just assigned them different colours using a computer program.
OK, so networks might look pretty, but how do they help us understand what's going on inside a cell? Well, for one thing, we know that a cell has lots of different things to do: producing energy to keep going, manufacturing proteins, sending signals to other cells and so on. Many of these things depend on what type of cell it is: a blood cell might need to carry oxygen, whilst a brain cell might need to process incoming signals (such as a signal for RED from the eyes) and send out a response (such as something corresponding to 'FIRE!' or 'DANGER!'). But whatever the type of cell, one might expect that there are certain kinds of proteins that specialise at certain kinds of tasks; and the proteins doing a particular thing probably tend to 'stick together', i.e., interact mostly amongst themselves. So the communities in the network above might correspond to groups of proteins that are involved in carrying out a particular task. Indeed, we do find that this is often the case: for example, some of the communities in the picture above are groups of proteins that make up the ribosome, the factory that manufactures new proteins. So by looking at where a protein sits in the network, and what other proteins it interacts with, we can usually get a pretty good idea of what the protein must be doing.
So far, we've been thinking of individual cells as somewhat static, boring creatures: unceasingly, unchangingly doing their job, whether it be carrying oxygen or stretching and contracting. But the fact is that cells keep changing; they have a life-cycle, just like us, and they go through different stages: growing, responding to the environment, dividing into daughter cells, wearing out, dying. Often it is the way in which these changes happen that we are particularly interested in understanding; for instance, cancer happens when for some reason the growing and dividing stages go into overdrive. From the protein network point-of-view, what is happening as the cell goes through these different stages? One way to think of it is that at each stage, only part of the network is 'switched on'; the cell is making only those proteins it needs for that stage. So the network shown above is not really static, but dynamic: imagine different parts of it lighting up at different times. If we take this into account, can it help us to better understand what roles the different actors (proteins) are playing in the great cellular drama? One interesting idea that was suggested by scientists some years ago was that if we focus on the seemingly important proteins, the ones that have many interactions (these are called 'hub' proteins), maybe by looking at when these interactions light up we can say something about what kind of protein it is.
Supposing I am a hub protein in the network, and I have five 'partners', proteins that I can attach to. One might imagine two opposing scenarios: maybe all my partners get produced by the cell at the same time, and so all the interactions get switched on at once. In this case, it's like me and my 5 partners all coming together for a big party; hence according to the scientists, I would be called a 'party hub'. On the other hand, it could be that my 5 partners get switched on by the cell at different times, as they are needed in different life stages. In this case, my interactions don't happen all at once, but one by one: they're like going on a sequence of dates with different partners, and so I would be called a 'date hub'.
 |
The idea that hub proteins came in two flavours, date and party, was quite exciting to scientists: the article that first came up with it was published in Nature, perhaps the most prestigious scientific journal. The reason for this was that the two types of hubs appeared to have distinct, important roles in how the protein network as a whole was organised. It appeared that the party hubs were like local, low-level coordinators: they helped to bring together many proteins that all had the same purpose, and were thus produced at the same time. For example, a party hub might attach to many other proteins to form a big protein factory, or ribosome. On the other hand, date hubs looked more like high-level, global organisers; they could help the different parts and stages of the network to communicate with each other, by for instance transmitting signals from one type of protein to another. A knowledge of what specifically date and party hubs were doing could be a major step forward in our goal of understanding how the complicated protein cocktail produces specific kinds of cells and behaviours.
Unfortunately, things turn out to be not so simple. Several other scientists disputed the idea that the hub proteins could be categorised into 'date' and 'party' types, presenting evidence that there was no consistent relationship between the pattern in which the interactions light up and the sort of role the protein has in organising the network. In our recent article in the journal PLoS Computational Biology, we establish this more clearly, showing that the so-called date hubs are not really any more likely to be global network coordinators than the party hubs. Moreover, protein hubs seem to display a wide variety of 'lighting up' patterns for their interactions, and classifying the hubs into just two types appears too simplistic.
It is not all bad news, however. So far, we have been thinking of individual proteins as the actors to whom we want to assign roles, in order to see how they fit into the bigger picture. But what if we instead think of interactions between proteins as the actors? In other words, what if we try to assign roles to the lines in the networks pictured above, rather than the dots? What kind of role could a line, or link, in the network have? Well, one way of thinking of it is to imagine that the links are roads, joining up a bunch of cities (the dots). In this case, if I want to drive from one city to another, I will naturally try to find the shortest path, i.e., the one with the smallest number of links or roads (let us say that all the roads have the same length). For any given pair of cities, I can come up with a shortest path between them. Now, suppose I remove one of the links; one road suddenly gets destroyed. How many of those shortest paths between cities have to be re-routed? If the answer is lots, then it means that the link we removed was very important to efficiently connecting up different points in the network. So, for each road or link, we can say that one way of measuring its importance is how many paths have to be re-rerouted if that link gets removed: this is technically called the betweenness of the link.
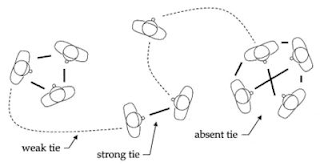
How is betweenness relevant to the network of proteins? What we found was that there is a strong relationship between how important a link is (in the sense just described), and how similar the two proteins joined by that link are: the links with the highest betweenness tend to be interactions joining the most dissimilar proteins. Hang on a minute, you may be thinking: how do I decide how 'similar' two proteins are? Well, we know quite a lot about proteins from experiments: for many of them we know about their structures, and about what parts of the cell they are found in, and about what sorts of tasks they seem to be involved in. We can use a database of such information to measure how much two proteins match up in these terms. Our results seem fairly consistent across different protein networks. So why is this interesting? One reason is that it seems to mirror something sociologists have long known about social networks (which are made up of people joined by relations like friendship or kinship; Facebook is just a giant online social network). In social networks, there is often a distinction made between 'weak' ties (or links) and 'strong' ties. Strong ties are close relations or friends; weak ties may be colleagues or acquaintances with whom one has less frequent interaction.
The interesting thing is that the weak ties turn out to be the most important ones for communicating information across the network: for example, if you are looking for a job, then it is more likely that someone outside your immediate social circle (say, a friend's colleague) will be able to provide a useful tip than someone whom you know very well. This simply reflects the fact that your nearest and dearest are most likely to share your own connections and information sources, whereas an outsider is more likely to know something novel. Coming back to protein networks, if we think of the betweenness of a link as a way of measuring its importance for information flows between proteins, then we see that here too the most important or central links are 'weak', in the sense that they are between dissimilar proteins that have different functions and are not part of the same group. This suggests that a deeper understanding of the roles played by specific links in protein networks, along the lines of things like weak and strong ties, may help us to unravel the tangled webs of proteins that comprise and control cells, and thus ultimately, life itself.